
Here we offer surface topography analysis for full-length PDB/AF2 proteins. Functional pockets are predominantly located on the protein surfaces and can also appear as buried cavities, both are analyzed by CASTpFold. Among structures, the vast number of 214 million AF2-predicted structures (as of Nov 2022) can be grouped into 18,661,407 clusters, whose representative structures are recognized by Foldseek (Inigo, et al. 2023). Among sequences, removing all sequences labeled as "fragment" in the Uniprot database, there are 2,302,908 non-singleton clusters remaining(as of Dec 2023), which can be mapped to 183,581,108 AF2-predicted structures. CASTpFold provides detailed surface topography analysis for these 2,302,908 non-singleton exempary structures, as well as the PDB structures.


Beyond direct matching of PDB/AF2 representative structures, querying an AF2 protein against the pool of 183,581,108 structures is automatically routed to its corresponding representative AF2 structure, as depicted in Fig A. When a search of an AF2 identifier is not found by CASTpFold, it is likely the query protein is not a full sequence but a "fragment". The search result for such cases is illustrated in Fig B.
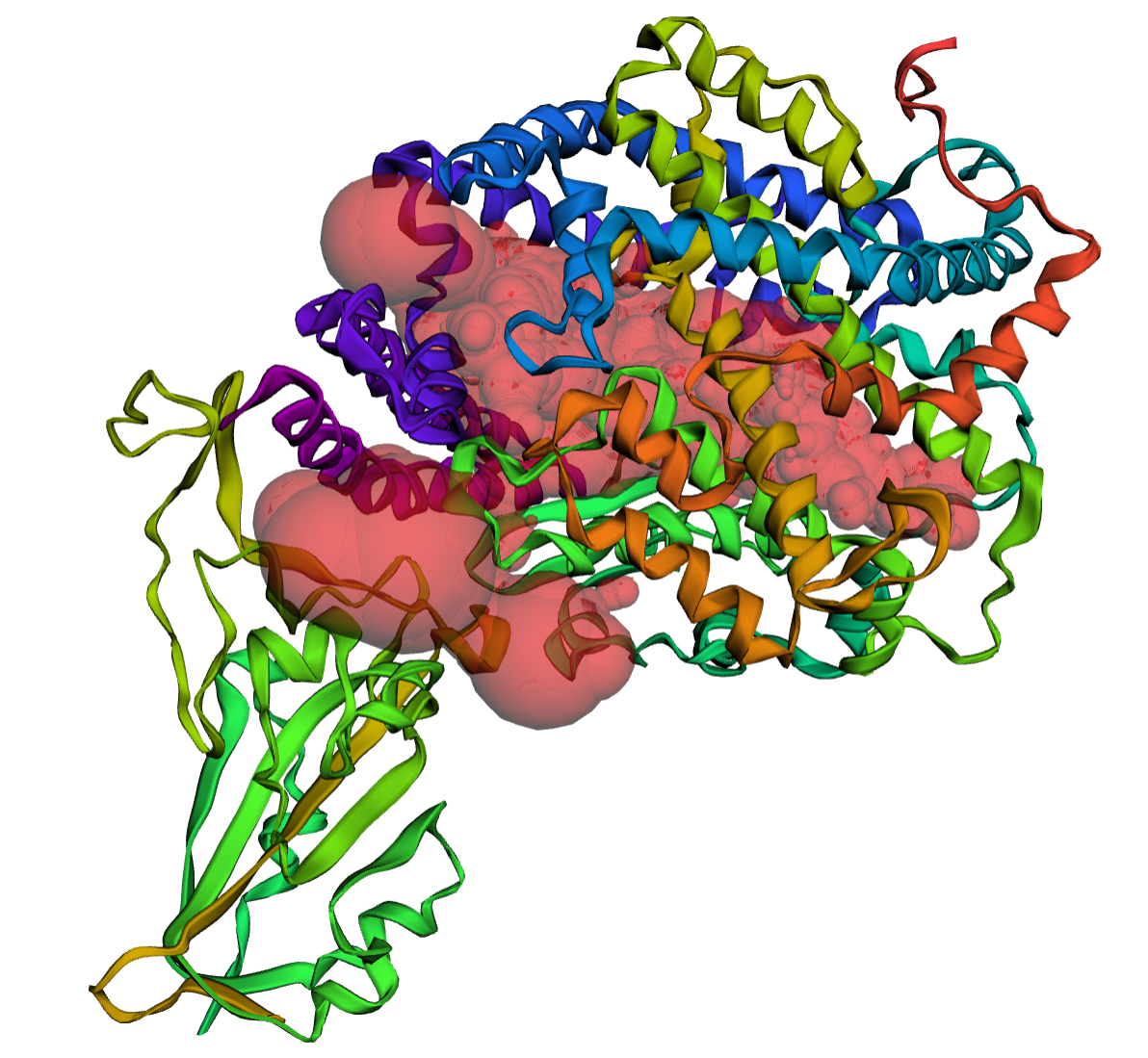
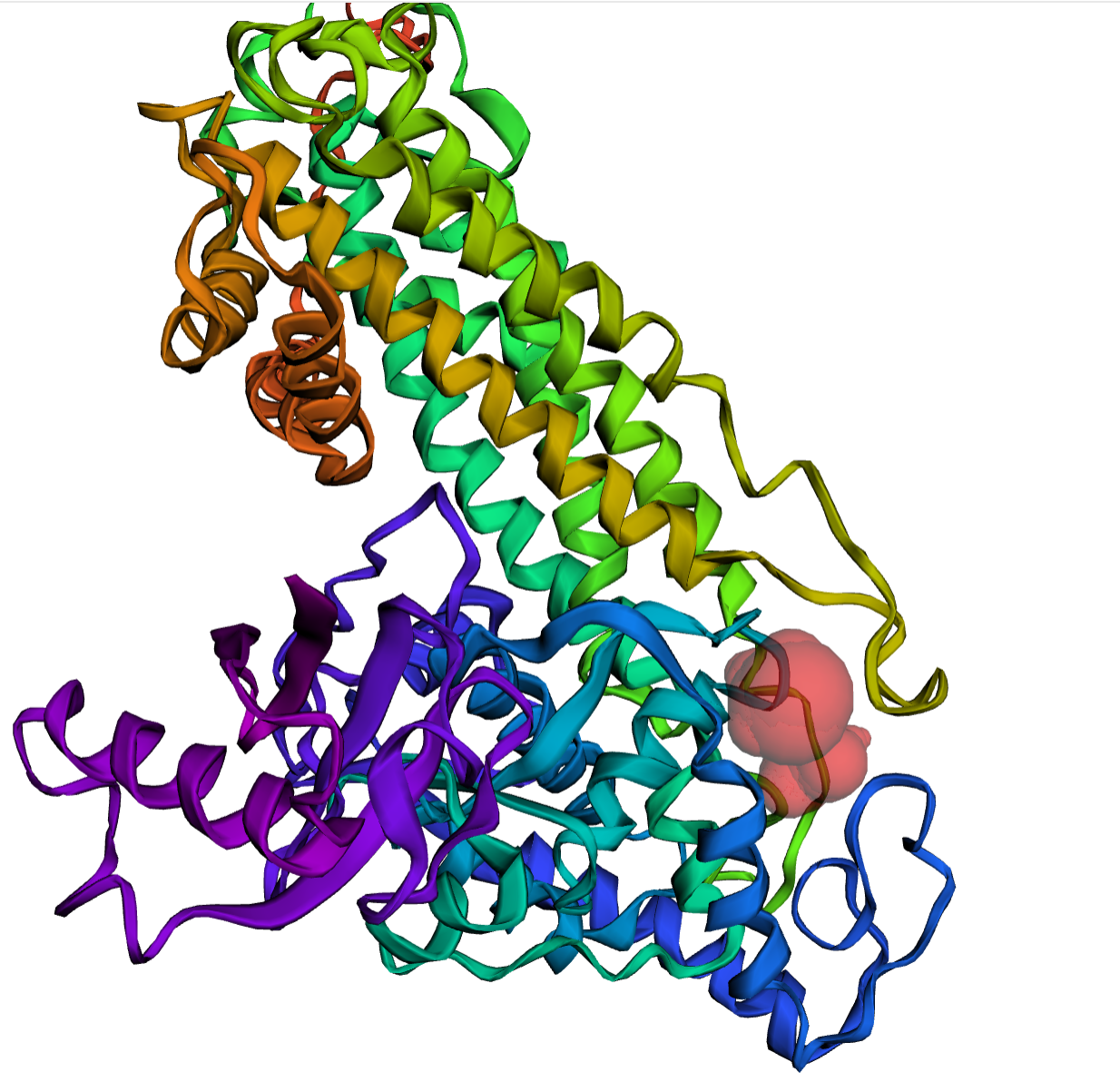
In our dataset of PDB structures and 2.3 million AF2-predicted representative structures, we have collected 3,684,958 surface pockets and 852,464 protein-protein interface (PPI) pockets. Each pocket contains at least 14 residues.
PPI pockets are regions where multiple protein chains interact to form protein complexes. They play important roles in recognition and binding among proteins. In Fig A, we show an example of a PPI pocket highlighted in red volume, which exists between the Sars-Cov2 Spike protein and the Human ACE2 protein. Surface pockets are regions found on the surface of a single protein chain. They can serve as active sites for catalytic reactions or as binding sites for molecules like substrates, cofactors, or regulators. In Fig B, a red volume is used to represent an active site-like pocket on the AF2-predicted human histidine ammonia-lyase structure.
We have clustered both databases of the PPI pockets and the surface pockets, and have identified groups of similar pockets within each dataset. We used the Foldseek algorithm for this task (Michel, et al, 2023). For all clusters of PPI and surface pockets containing a minimum of 14 residues from a single protein/complex, users can access and download the list of similar pockets and visualize them in the "Pocket Similarity" section.
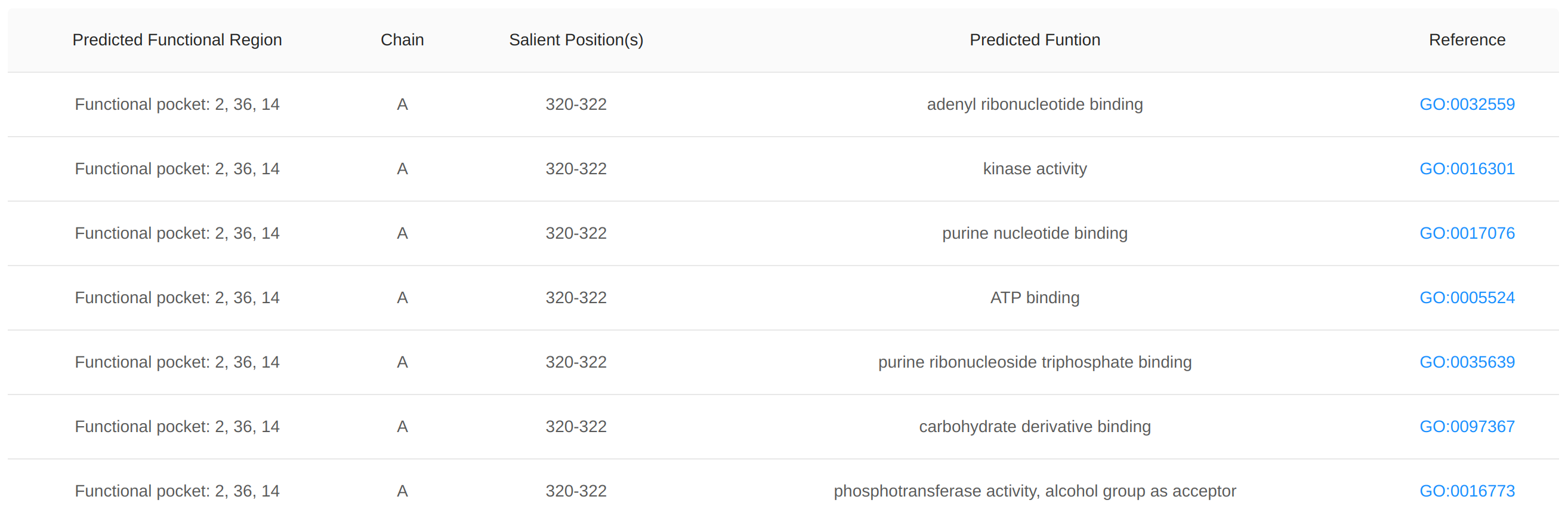
We use the DeepFri method to predict functions of proteins in GO terms/EC numbers (Vladimir, et al. 2021), with the relevant residues identified, for all 2.3 million AF2-predicted representatives. Additionally, we pinpoint the functional pocket involved in carrying out these functions, where the identified function-related residues are located. An individual pocket may be predicted to serve multiple functions, as illustrated in the figure with the example for AF2 ID: A0A178E137.
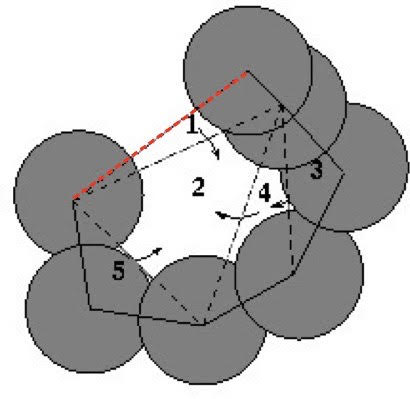
After detecting the pocket based on the alpha-complex (see background), we identify atoms forming "rims" of the pocket mouth(s). A two-dimensional mouth is the red dotted edge on the boundary of the pocket (upper edge of triangle 1) in Fig A, and a three-dimensional pocket (PDB: 2iwv) has two mouths from the top and bottom shown in Fig B.
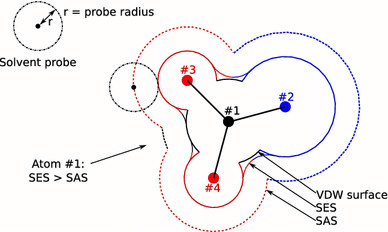
The difference between Solvent Accessible and Solvent Excluded or Molecular Surface model (MS) based surface can be found in Fig (Diogo, et al, 2016). In CASTpFold, we only show the SA surface area and volume (Liang et al, 1998b). The pocket solvent accessible surface area (SASA) is the two-dimensional measure describing the SA surface area of a pocket that is accessible to solvent molecules, and the pocket solvent accessible volume (SAV) is the three-dimensional measure that refers to the volume filling the pocket that is accessible to solvent molecules. Here area is measured in Å2, and volume in Å3.